Abstract
Background: Atopic
diseases have a strong genetic predisposition. Many attempts
have been made to identify the genes related to atopy. A
mutation in the human interleukin -4 receptor
α ( hu
IL-4Rα)
has been linked with atopy in human.
Objective: We examined
the relationship between the variation at amino acid 551 of
hu IL-4Rα and atopic dermatitis
(AD).
Methods:
PCR- based
restriction fragment length polymorphism assay was used to
investigate the relationship between the glutamine 551
Arginine (Gln551Arg) and AD. Furthermore the serum IL-4 and
total IgE levels were measured by using ELISA technique.
Results: The
prevalence of Arg551 in AD patients was significantly higher
than in controls (p<0.001). Relative risk of AD among those
with a mutant allele Arg551 is 7.36. Arg551 allele was
significantly correlated with the disease severity (p<0.05)
and with the serum total IgE level (p<0.01) but not with the
serum IL-4 level (p>0.05).
Conclusion: The Arg551
allele of IL-4Rα is strongly
associated with AD and may serve as a clinically useful
marker of AD severity.
Introduction
Atopy is complex, multifactorial disorders characterized by
the formation of IgE antibody in persons with a genetic
predisposition. A number of atopy susceptibility genes have
been identified. The difficulties of conducting genetic
studies of atopy are due to in part the multiple genetic
markers for atopy [1].
Interleukin-4 is a peptide secreted by TH2 cells
and mast cells; it is the major cytokine
responsible for the induction of IgE synthesis by B cells.
Furthermore, IL-4 acts on TH0 cells and promotes
their progression to the TH2 phenotype; ultimately
leading to the secretion of more IL-4 and other TH2-derived
cytokines. It also induces the adhesion molecules on
endothelial cells which important for eosinophil migration
thereby perpetuating the allergic cascade [2].
The IL-4 receptor is composed of two subunits; a 140-kd α-subunit,
which binds IL-4 and transduces its growth-promoting and
transcription –activating functions, and a γ
chain subunit which amplifies signaling of IL-4Rα.
[3]. Signal
transduction of the receptor requires phosphorylation by
intracellular kinases which phosphorylate some or all of the
five conserved tyrosine (Y) residues in the cytoplasmic tail
of IL-4Rα. [4].
An IL-4Rα allele was identified in
which guanine was substituted for adenine at nucleotide 1902,
causing a change from glutamine to arginine at position551
(numbering from the start of the mature protein) (Arg 551) in
the cytoplasmic domain of the IL-4Rα
protein. Arg 551 has been reported to be correlated with
hyper IgE syndrome and severe atopic eczema, and occurs with
a frequency of 20% in the general population. Individuals who
possess 1 or 2 copies of this allele have significantly
increased relative risk toward the atopic phenotype compared
with the wild-type variants with glutamine at position 551
(Gln 551)[5-6].
This work had been designed for
identification of the allelic distribution of the Gln551Arg
polymorphism in AD and its usefulness as a clinical marker of
the disease. Also, study the relation of the allele with the
atopic markers (serum IL-4 and serum total IgE) and with the
disease severity.
Subjects and Methods:
This study included 20
patients with atopic dermatitis of both sexes with an
age ranging from 8-22 years and 10 normal control subjects
(age and sex matched). All patients met the diagnostic
criteria for atopic dermatitis, as defined by Hanifin
and Rajka [7] None of these
patients had other atopic conditions (asthma, rhinitis or
conjunctivitis) nor received antihistamines, or systemic or
topical corticosteroids during the period of 3 weeks before
clinical evaluation. The severity of atopic dermatitis was
measured by using the SCORAD index [8].
AD was considered mild ,moderate, and severe forms in which
the SCORAD index was less than 25, between 25 and 50 and 50
respectively.
Both patients and healthy
control were subjected for the following assessment:
-Genotyping for Arg551
and Gln551 IL-4Rα alleles by
PCR-based restriction fragment length polymorphism assay
according to Rosa-Rosa et al [6]
.
-Serum IL4 level was
measured by ELISA using the Bio-Source International, Inc.
human IL-4 (hIL-4) kit according to Banchereau
[9].
-Serum total IgE level
was measured by ELISA using the Elitech IgE quantitative
according to Dorrington& Bennich [10].
Genetic analysis
Genomic DNA isolated from 300 µL
whole EDTA blood with the use of the PUREGENE DNA Isolation
Kit purchased from Gentra (Minneapolis) was analyzed for
the presence of the Arg 551 or Gln 551 alleles by PCR.
PCR
was done by using the nested primers (Biosource Europe S.A.,
Belgium, Netherlands German) 5' - TCT CGG CCC CCA CCA GTG
GCG ATC - 3' (sense) and 5' - GAG GTC TTG GAA
GAG CUT
ATA C - 3' (antisense) and was carried out in a total volume of
16.44µL under the following conditions : 1 cycle of 94 °C for
3 minutes, 32 cycles of 94 °C for 20 sec.,58 °C for 30 sec. and 72
°C for 30 sec., 1 cycle of 72 °C for 5 minutes
and one cycle of 4 °C. Restriction digest reaction was
performed in a total volume 30µl with 5 units of PvuI (Proteous
Vulgaris-I) (Amersham Place Little Chalfont Buckinghamshire
England HP7 9Na). The digested products were fractionated in
an agarose gel matrix using the EC 360 Submarine Gel
electrophoresis system (Maxicell, EC 360 M-E-C apparatus
Cooperation St. Petersburg. Florida USA).The Gln551 and
Arg551 alleles yielded 209bp and 186bp bands respectively.
Statistical analysis
Data were expressed as range, mean ± SD. ANOVA test,
student -t-test, Fisher's exact test and correlation
co-efficient (r) were used to test the significance of the
results. Non parametric data was analyzed by Chi Square test.
Relative risk (RR) is defined as the probability of an event
of the active group divided by the probability of the event
in the control group. P<0.05 was accepted as statistically
significant.
Results
Table (1)
shows the demographic data for the study groups. As regard
age and sex there were no statistically significant
differences between the AD and control groups (p > 0.05).
PCR-based Restriction Fragment
Length Polymorphism assay (RFLP) of Gln551 and Arg551 alleles
revealed a 209-bp band from Gln551 allele and a 186–bp band
from the Arg551 allele
(Figure 1).
|
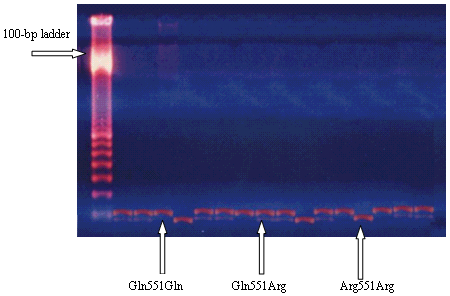 |
Figure 1: PCR-based RFLP assay.
Arg551 and Gln551 IL-4Rα alleles correspond to 186- and 209- bp bands, respectively.
|
In control group, 8 persons had
homozygous Gln551Gln (80%) and 2 persons (20%) had
heterozygous Gln551Arg, and none had homozygous Arg551Arg .
The Arg551 allele was
found in only 10% (2/20) , the allele frequency for the
Gln551 in 90% (18/20). Of the 20 AD patients, the homozygous
Arg551Arg was found in 4 patients (20%), the heterozygous
Gln551Arg in 10 patients (50%), and the homozygous Gln551Gln
in 6 patients (30%). The allele frequency for Arg551 allele
in AD group was 45% (18/40). There was a highly significant
association of Arg551 allele in AD than in control group (p
< 0.001). The allele frequency for the Gln551 in AD group was
55% (22/40). There was a highly significant association of
Gln551 allele in control group compared with AD group (p
< 0.001). There was also a significant association of
homozygous Arg551Arg in AD group over the control group (p
< 0.05). The relative risk for Arg551 allele in AD was
7.36. These results are shown in
Table 2.
Of the homozygous Gln551Gln AD
patients 4/6 (66.7%) had mild dermatitis, 2/6 (33.3%) had
moderate dermatitis, none had severe dermatitis. There was a
highly significant association of this allele with mild
versus severe dermatitis (p<0.05, exact Fisher test).
Of the homozygous Arg551Arg AD patients; severe dermatitis
was found in 2/4 patients (50%), moderate dermatitis was
found in 2 patients (50%), and none had mild dermatitis.
These results were statistically significant for severe
versus mild dermatitis (p < 0.05). Of the heterozygous
Gln551Arg AD patients, 4/10 patients (40%) had mild
dermatitis, 4/10 patients (40%) had moderate dermatitis, and
2/10 patients (20 %) had severe disease (p > 0.05),
Table 3.
Table 4 shows the
difference in serum IL-4 and serum total IgE levels between
patients and control group. There was a highly significant
increase of serum IL-4 and serum total IgE in AD group (p <
0.001).
The difference in serum IL-4
level in AD patients according to the severity of the disease
and different alleles was not significant (p>0.05), however,
in serum total IgE the difference was significant according
to the severity of the disease and different alleles (p<0.001
and p<0.01 respectively) (Table 5&6) .There was no
statistically significant correlation between serum IL-4
values and the SCORAD in AD patients (r = 0.243, p=
0.303)(Fig.2). There was a highly significant positive
correlation between serum total IgE and the SCORAD in AD
patients (r = 0. 924, p < 0.001) (Fig. 3).
|
Table (1): Demographic data
in the studied groups. |
| |
Control (N=10)
|
AD patients (N=20)
|
Age (yrs) Mean± SD
|
15.2±5.45
|
13.4±4.30
|
t
P |
0.987
>0.05 NS
|
|
Sex |
No. |
% |
No. |
% |
Male
Female |
7
3 |
70
30 |
12
8 |
60
40 |
X2
P |
0.17
>0.05 NS |
|
Table (2): Frequency of Arg551 and Gln551 IL-4Rα alleles
in atopic dermatitis versus control group. |
Population |
Gln551Gln |
Gln551Arg |
Arg551Arg |
Atopic dermatitis (n=20)
No.
%
Alleles (n=40)
No.
Allele frequency (%)
|
6
30
Gln551
22
55
|
10
50
|
4*
20
Arg551
18
45** |
Control group (n=10)
No.
%
Alleles (n=20)
No.
Allele frequency (%) |
8
80
Gln551
18
90 |
2
20
|
0
0
Arg551
2
10 |
Relative risk for Arg551
allele and atopic dermatitis is 7.36.
*p value for Arg551Arg is 0.03 between atopic dermatitis
patients and controls (Exact Fisher test).
**p value for Arg551 allele is < 0.001 between atopic
dermatitis patients and controls (chi square test).
|
|
Table (3): Effect of Arg551
and Gln551 IL-4Rα allelic variant on atopic dermatitis
severity. |
Dermatitis severity
|
Gln551Gln |
Gln551Arg |
Arg551Arg |
Mild
Moderate
Severe
p
value: severe
vs
mild |
4 (66.7%)
2 (33.3%)
0 (0%)
0.02
|
4 (40%)
4 (40%)
2 (20%)
0.69
|
0 (0%)
2 (50%)
2 (50%)
0.03
|
Mild: SCORAD < 25.
Moderate: SCORAD 25-50.
Severe: SCORAD >50
|
|
Table (4): Serum IL-4
levels (pg/mL) and total IgE levels (IU/mL) in the studied groups |
| |
Control
(N=10) |
AD patients
(N=20) |
|
IL4
range
Mean± SD |
(13.9-38.23)
27.06±7.27 |
(55.28-81.97)
68.69±7.44 |
|
t
P |
14.558***
<0.001 HS |
|
|
IgE
range
Mean± SD |
59.75-149.1
102.12±30.59 |
192.3-479.2
296.45±80.9 |
|
t
P |
7.286***
<0.001 HS |
|
|
Table (5): Serum IL-4 (pg/mL), and total IgE (IU/mL)
in AD patients according to the severity of the disease |
| |
Mild (no.=8) |
Moderate (no.=8) |
Sever (no.=4) |
F |
P |
IL-4
|
67±7.7
|
69.23±8.24
|
77.7±14.13
|
0.801
|
> 0.05 NS
|
IgE
|
225.27±19.54
|
306.6±42.51
|
418.47±51.99
|
36.596***
|
<0.001HS
|
F: ANOVA test which compares
the means of several groups.
|
|
Table(6): Serum IL-4 (pg/mL),
Serum total IgE (IU/mL) and different alleles in atopic
dermatitis group (n= 20). |
| |
Gln551Gln
(N=6)
|
Gln551Arg
(N=10)
|
Arg551Arg
(N=4)
|
F |
P |
IL-4
|
66.4±8.6
|
70.6±8
|
67.2±2.48
|
0.675
|
> 0.05 NS
|
IgE
|
243.36±48.77
|
278.05±66.92
|
396.57±70.25
|
7.597**
|
<0.01S
|
F: ANOVA test which compares
the means of several groups.
|
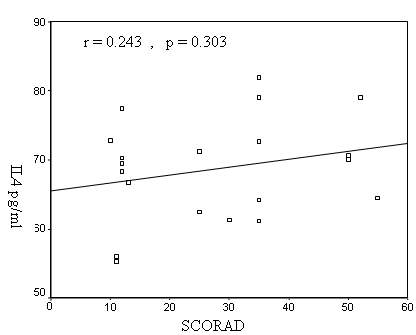
Figure 2: Correlation between serum IL-4 and SCORAD in
AD patients
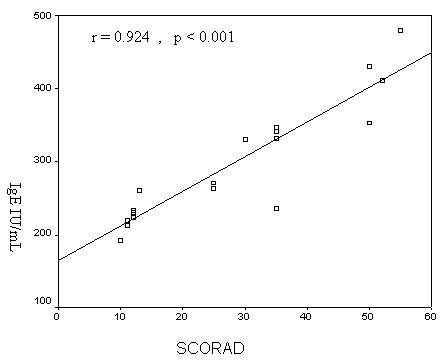
Figure 3: Correlation between serum total
IgE and SCORAD in AD patients.
Discussion
It is
increasingly evident that many different genes influence the
development of atopy. Three different classes of
disease-associated genes have been described: susceptibility
genes that are strongly associated with atopy and likely play
a causative role, disease-modifying genes that are not
causative but modify the phenotype of the disease, and
drug-modifying genes that alter the response to
pharmacologic agents in affected individuals [6].
In current study,
Arg551 allele acts as an atopy
susceptibility gene, as the allele frequency for Arg551
allele in this study was 45% in AD group compared with 10% in
the control group. Thus individual carries Arg551 allele has
7.36–fold enhanced risk toward AD than does the individual
with Gln551 allele. Furthermore, there was a strong
association of the homozygosity of Arg551 allele and AD, as
20% of our AD patients were homozygous while none of the
control was homozygous for the Arg551 allele.
In
agreement with the present study, some authors [11-13]
reported that the Gln 551Arg allele frequency was
more common in patients with AD than the controls, while
these results conflict with others [14-17].
The
suggested molecular mechanism underlying the observed
enhanced signaling with Gln551Arg mutation and the
association with Arg551 allele with atopy is that the
substitution of arginine for glutamine at position 551(numbering
from the start of the mature protein)
alters the binding profile of the
adjacent phosphorylated tyrosine residue (Y550) and decreases
the binding of phospho-tyrosine phosphatases (SHP-1). SHP-1
dephosphorylates regulatory phospho-tyrosine residues and has
been implicated in the termination of IL-4 receptor signaling
leading to exaggerated IL-4 responses. Alterations
in the dephosphorylation of signal transducers and activators
of transcription (STAT-6) should have a potent effect on
IL-4-mediated responses [5,18].
This explanation coincided with the recommendation of
Kamata et al, [19] who reported
that reduced SHP-1 protein expression results in enhanced
IL-4R–mediated signal transduction and TH2
cytokine production.
However, Wang et al, [20]
noted that Arg551 allele does not have a direct effect on
IL-4 signal transduction. It is possible that multiple
docking sites for such a phosphatase exist on the huIL-4Rα so
that altering a single site would not result in a dramatic
change in signaling. It is possible that for an effect of the
Arg551 allele change to be observed, an additional mutation
in the huIL-4Rα or a mutation in, or amplification of one of
its signaling molecules including Janus tyrosine kinase
(JAK1,JAK3) and STAT-6 must also occur.
Results of the present study revealed a significant
association of the homozygosity of Arg551 allele and AD
severity (50% of homozygous Arg551 of patients had severe
dermatitis) thus, in agreement with previous studies it can
be postulated that Arg551 allele acts as a disease-modifier.
[5,6,21,22,23].
A majority of subjects identified as carrying a
single copy of the mutant allele were found to have atopy,
suggesting an intermediate dominant effect, with
(increasing) homozygous suffering more severe form, and
heterozygous more likely to develop more mild form of the
diseases (gene dosage effect). However, the finding that some
carriers of the Arg551 allele, including one who was
homozygous, were not atopic indicates that the penetrance of
this allele may be modified by other factors. These may
include distinct genetic loci that impart susceptibility to
or protection from atopy and environmental factors such as
the level and duration of exposure to allergens [24].
This mode of inheritance conflicts with the French
group [25], who reported that
the Gln551Arg allele was significantly more common among
atopic subjects and seemed to act as a recessive.
The inconsistent findings in the genetic studies of
atopy may partly be explained by ethnic differences in nature
and frequencies of genetic variants in disease susceptibility
genes. It can be also speculated that ethnic differences in
inflammatory genes (in addition to environmental factors) may
also underlie the significant worldwide differences in the
prevalence of AD. So, the controversial results may be due to
interaction between the mutation and the environment. This
suggestion is supported by Callard et al, [26]
who have found that there is an association between the
Arg551 polymorphism and flexural eczema in children at 6
months of age who have not had infection requiring treatment
with antibiotics. Antibiotics supporting the
''hygiene hypothesis'' , which states that exposure to
infection in childhood can protect against allergic disease .
Liu et al, [27] gave
significant evidence supporting the interaction between
exposure to maternal smoking and variant Gln551Arg on risk of
cat sensitization.
These controversial results pointed to the importance of
determining the frequencies of single nucleotide
polymorphisms in different populations before drawing
conclusions from allele association studies, since the
background allele frequencies may be disparate between
different populations.
In the present study the serum level of IL-4 was
significantly higher in AD patients group than control group
(P < 0.001). no significant difference was found in its level
between Arg551Arg, Gln551Arg, Gln551Gln groups (p>0.05). It
was not correlated with the SCORAD (r =0.243, p>0.05). This
indicates, that serum level of IL-4 is an indicator of the
presence of atopy not the disease severity. Previous
studies [28,29]
have also reported a significant increase in serum level of
IL-4 in AD patients than controls.
Grewe et al, [30]
tried to explain the absence of correlation of IL-4 with
the atopic disease severity; the authors stated that IFN-γ
but not IL-4 has been correlated with the clinical severity
of AD. This may be related to the capacity of IFN-γ to
enhance eosinophil viability, augment eosinophil activation
and cytotoxic activity, activate vascular endothelial
molecules, which increase the infiltration of eosinophils,
thereby contributing to AD. This is supported by Ochiai et
al,
[31]
who found inverse correlation between
IFN-γ:IL-4 ratio. So, the balance between TH1
(IFN-γ) and TH2 (IL-4) cytokine production is
important, rather than the absolute levels of TH2
cytokines, that decides clinical severity.
In the present study, there was a highly
significant increase in serum total IgE in AD group as
compared with the control group (P<0.001).Also, there was a
highly significant increase in serum IgE levels in homozygous
Arg551Arg, than heterozygous Gln551Arg, than the homozygous
Gln551Gln (p<0.01). A positive correlation was found between
increased serum total IgE levels and SCORAD(r=0.924,
p<0.001). These results indicate that serum total IgE may be
used as indicator of AD and its severity. Significant higher
levels of serum IgE have been found in other studies [32,33]
.
An association between Arg551 allele and
increased IgE level in AD was reported in many studies [5,6,34],
which support the suggestion that Arg551 allele confers
genetic susceptibility to atopy. On the other hand Kruse
et al, [35] have found an
association of Arg551 allele with lowered serum total IgE
concentrations.
Some
authors [32,36]
have reported a significant correlation between
dermatitis severity and serum total IgE levels, while
Wolf [37] has found
that 20% of the patients with severe atopic disease have
normal or subnormal serum total IgE levels, and that the
severity of the disease does not always correlate with serum
total IgE levels. These findings do not reduce the importance
of this antibody in the onset of the illness. A possibility
is suggested here that even low IgE concentrations are
capable of playing a key role in the pathogenesis of the
disease, and being directly responsible for its clinical
manifestations.
In
conclusion, the Arg551 allele is an atopic
dermatitis susceptibility gene as well as a
disease-modifying gene that may be a useful genetic marker of
the disease severity.
References
1- Mackay,
I.R., and Rosen, F.S.: Advances in immunology; Allergy
and allergic diseases. N. Engl. J. Med. 343: 30-37,2001.
2- Lordan, J.L.; Bucchieri,
F.; Richter, A., et al.: Cooperative effects
of Th2 cytokines and allergen on normal and asthmatic
bronchial epithelial cells. Immunology. 169:
407-414, 2002.
3- Kawakami K., Taguchi J.,
Murata T., et al.:
The interleukin -13 receptor
alpha-2 chain: an essential component for binding and
internalization but not for interleukin-13 induced signal
transduction through the STAT6 pathway. Blood 97:2673-2679,
2001.
4- Nelms K., Keegan A.D., Zamorano J., et al.:IL-4
receptor: signaling mechanisms and biological functions.
Annu.Rev. Immunol.,17:701-738,1999.
5- Hershey G.K.K.,
Friederich M.F., Esswein L.A. et al.: The
association of atopy with a gain function mutation in the
α-subunit of the interleukin-4 receptor. N. Engl. J. Med.
337:1720-1725,1997.
6- Rosa-Rosa, L.; Zimmermann
N,; Bernstein, J.A.; et al.: The R 576 IL-4 receptor α
allele correlates with asthma severity. J. Allergy Clin.
Immunol. 104:1008-1014,1999.
7- Hanifin, J.M. and
Rajka, G.: Diagnostic features of atopic eczyma. Acta.
Dermatol. Venareol. 92:44-47,1980.
8- Severity
Scoring of atopic dermatitis: the SCORAD index. Consensus
report of the European Task Force on atopic dermatitis. Dermatology , 186:
23-31,1993.
9- Banchereau, J.:
Interleukin-4. Medecine/Science. 6:946-953,1990.
10- Dorrington, K.L. and Bennich, H.H.: Structure-Function Relationship in
Human Immunoglobulin-E. Immunol. Rev.41:3-25,1978.
11-
Dupre, D.; Audrezet, M.P.
and
Ferec, C.:
Atopy and a mutation in the interleukin-4 receptor gene.
N. Engl. J. Med. 343:69-70,2000.
12- Oiso, N.; Fukai, K.
and Ishii, M.: Interleukin 4 receptor alpha chain
polymorphism Gln551Arg is associated with adult atopic
dermatitis in Japan. Br. J. Dermatol. 142:1003-1006,2000.
13- Yandava C.N.; Pillari A.;
Lilly C.M. et al.: An association of interleukin-4 alpha
receptor gene mutation and asthma and atopy. Am. J. Hum.
Gen . 63:89,2005.
14- Mitsuyasu, H.;
Yanagihara, Y.; Mao, X.Q.; et al.: Cutting Edge:
Dominant Effect of Ile50Val Variant of the Human IL-4
Receptor {alpha}-Chain in IgE Synthesis. Immunology. 162:
1227-1231,1999.
15-
Tan, E.C.; Lee, B.W.; Tay, A.W.; et al.:
Interleukin-4 receptor variant Q576R: ethnic differences and
association with atopy.
Clin. Genet. 56:333-334,
1999.
16- Patuzzo, C.; Trabetti,
E.; Malerba, G.; et al.: No linkage or association of
the IL-4Rα gene Q576R mutation with atopic asthma in Italian
families. J. Med. Genet. 37:382-384,2000.
17- Haagerup, A.; Bjerke, T.;
Schiotz, P.O.; et al.: No linkage and association of
atopy to chromosome 16 including the interleukin-4 receptor
gene. Allergy. 56:775-779,2001.
18- Hanson, E.M.;
Dickensheets, H.; Qu, K.; et al.: Regulation of the
Dephosphorylation of STAT-6 participation of tyr-713 in the
interleukin-4 receptor, the tyrosine phosphatase shp-1, and
the proteasome. J. Biol. Chem. 278:3903-3911,2003.
19- Kamata, T.; Yamashita,
M.; Kimura, M.; et al. : Src homology 2
domain–containing tyrosine phosphatase SHP-1 controls the
development of allergic airway inflammation. J. Clin.
Invest. 111:109-119,2003.
20- Wang, H.Y.; Shelburne,
C.P.; Zamorano, J.; et al.:
Cutting Edge: Effects of an
allergy–associated mutation in the human IL-4Rα (Q576R) on
human IL-4 induced signal transduction. Immunology. 162:
4385-4389,1999.
21- Deichmann, K.A.;
Bardutzky, J.; Forster, J.; et al.: Common polymorphisms
in the coding part of the IL-4-receptor gene. Biochem.
Biophys Res Comm 231:696-697,1997.
22- Beghe, B.; Barton, S.;
Rorke, S.; et al.: Polymorphisms in the interleukin-4 and
interleukin-4 receptor alpha chain genes confer
susceptibility to asthma and atopy in a Caucasian
population. Clin. Exp. Allergy. 33:1111-1117,2003.
23- Hytonen, A.M.; Lowhagen,
O.; Arvidsson, M.; et al.: Haplotypes of the
interleukin-4 receptor alpha chain gene associate with
susceptibility to and severity of atopic asthma. Clin.
Exp. Allergy. 34:1570-1575,2004.
24- Peat, J.k.; Tovey, E.
and Toelle, B.G.: House dust mite allergens: a major
risk factor for childhood asthma in Austria. Am. J.
Respir. Crit. Care Med. 153:141-146,1996.
25- Ziani1, S.; Chavernoz1,
N.; Morgant1, M.; et al.: The Interleukin-4 receptor
variants I50V and Q576R in atopic children. Immunology.
162:541. 1999.
26- Callard, R.E.; Hamvas,
R.; Chatterton, C.; et al. : An interaction
between the
IL-4Ralpha gene and infection is associated with atopic
eczema in young children. Clin. Exp. Allergy.
32:990-993,2002.
27- Liu,
X.; Beaty, TH.;
Deindl, PH.; et al.: Associations between specific serum
IgE response and 6 variants within the genes IL4, IL13 and
IL4RA in German children. J. Allergy Clin. Immunol.
113:489-495,2004.
28- Kaminishi, K.; Soma, Y.;
Kawa, Y. et al.: Flow cytometric analysis of IL-4. IL-13
and IFN-gamma expression in peripheral blood mononuclear
cells and detection of circulating IL-13 in patients with
atopic dermatitis provide evidence for the involvement of
type 2 cytokines in the disease. J. Dermatol. Sci.
29:19-25,2002.
29- Nomura, I.; Goleva, E.;
Howell, M.D.; et al.:Cytokine milieu of atopic
dermatitis, as compared to psoriasis, skin prevents induction
of innate immune response genes. Immunology. 171:
3262-3269,2003.
30- Grewe, M.C.A.; Schopf,
B.E.; Thepen, T.; et al.: A role for Th1
and Th2 cells in
the immunopathogenesis of atopic dermatitis. Immunol.
Today 19:359-361,1998.
31- Ochiai, K.; Tanaba, E.;
Ishihara, C.; et al.: Role of JAK2 signal transduction
pathway in activation and survival of human peripheral
eosinophils by interferon- gamma (IFN-γ). Clin. Exp.
Immunol. 18:340-343 ,1999.
32- Kawai, K.; Kamei, N.
and Kishimoto, S.: Levels of serum IgE, serum
soluble-Fc epsilon RII, and Fc epsilon RII(+) peripheral
blood lymphocytes in atopic dermatitis. Dermatology.
19:285-292,1992.
33- Poljacki, M.; Jovanovic,
M. and Matic, M.: Immunologic response in patients
with atopic dermatitis. Med. Pregl. 49:305-307,1996.
34- Isidoro-Garcia, M.;
Davila, I.; Moreno, E.; et al: IL4RA gene polymorphism
(Q576R) is associated to higher total IgE levels in Spanish
patients with family history of atopy. Med. Clin. (Barc).
124:211-212,2005.
35- Kruse, S.; Japha, T.;
Tedner, M.; et al.: The polymorphisms S503P and Q576R in
the interleukin-4 receptor alpha gene are associated with
atopy and influence the signal transduction. Immunology.
96:365-371, 1999.
36- Laske, N. and
Niggemann, B.:
Does the severity of atopic dermatitis
correlate with serum IgE levels? Pediatr. Allergy Immunol.
15:86-88,2004.
37- Wolf, R.:
The role of
IgE in atopic diseases. Med. Hypotheses. 14:353-355,1984.
© 2006 Egyptian Dermatology
Online Journal
|

