|
|
Abstract:
Background: Dermatophytosis accounts for the majority of fungal
infection all over the world. The conventional laboratory methods for
identification of dermatophytes are slow and lack specificity. Genetic
amplification has made rapid and precise identification of dermatophytes
possible. With the increasing variety of drugs available for the treatment
of dermatophytosis and with the lack of effective and safe antifungal, the
need for a reference method for testing the antifungal susceptibilities of
dermatophytes has become apparent.
Aim of the study: The current study was conducted to compare the
rapid diagnostic molecular technique arbitrarily primed polymerase chain
reaction (AP-PCR) with the conventional culture method for identification of
the dermatophyte fungal infections of hair, nail and skin. Also to determine
the antifungal susceptibility pattern of different dermatophyte isolates to
Terbinafine, Griseofulvin, Itraconazole and Ketoconazole as the routinely
used antifungal agents. Patients and Methods: The present
study included 115 patients with dermatomycosis of the hair, skin and nail.
Their age ranged from 3 to 50 years (mean 19.8±12.5 SD). Specimens from the
infected sites were collected and subjected to conventional examination by
direct (potassium hydroxide) KOH microscopic examination, culture on primary
and selective media. Dermatophyte isolates were identified by their
characteristic morphology, physiological tests and AP-PCR. Antifungal
susceptibility was tested for all isolates according to the National
Committee for Clinical Laboratory Standards NCCLS microdilution method M38-A
for filamentous fungi with modifications in temperature and incubation
period. Results: Out of the 115 cases with ringworm
infection, dermatophytes were isolated in culture from 46.1% of specimens
and nondermatophytes from 18%. Trichophyton (T) rubrum (32.1%) was the most
commonly isolated dermatophyte from all types of skin fungal infection
except tinea capitis (P<0.001). T. mentagrophytes and T. violaceum were the
main causes of tinea capitis. By genotypic identification (AP-PCR) of
dermatophytes, all isolates formed distinct DNA band patterns on gel
electrophoresis which was in agreement with the conventional methods in
86.8% of isolates. Out of the eleven phenotypically identified T.
mentagrophytes; two only were diagnosed to the strain level, two strains
were genotypically identified as T. rubrum and one as T. tonsurans. Also two
isolates of T. violaceum were diagnosed by PCR as T. schoenleinii, one T.
rubrum was diagnosed as T. ajelloi and one T. soudanense as T. violaceum. The
direct KOH examination had sensitivity of 88% and specificity of 74%. The
antifungal susceptibility pattern of the isolated dermatophytes were for
terbinafine 0.06-0.5 (0.121) µg/ml, itraconazole 0.06-4 (0.62) µg/ml,
ketoconazole ranged from 0.06-4 (0.857) µg/ml and griseofulvin from 0.5-8
(2.151) µg/ml. Terbinafine was the most powerful antimycotic and T. rubrum
had the highest ( minimal inhibitory concentration) MIC values for the four
antifungal agents.
Conclusion: The genotypic differentiation by AP-PCR provides a
rapid and practical tool for identification of dermatophyte isolates to the
species and strain level within one day that is independent of the culture
variations. The standard NCCLS M38-A broth microdilution method with the
modifications in temperature and incubation period is convenient for
antifungal susceptibility testing of dermatophytes. Introduction
Dermatophytes infections of the skin affects a large proportion of
population and have emerged as important causes of morbidity especially in
aging population and in immunocompromised patients (Marie et al., 2001 [46]
and Osborne at al., 2003[59]). These dermatophytes are of
the genera Microsporum, Trichophyton and Epidermophyton. The genus
Epidermophyton is represented by a single pathogenic species (E. floccosum),
the genera Microsporum and Trichophyton are complex and made of multiple
species (Koichi et al., 1999)[38]. For
many years they were accustomed to diagnose dermatophytes on the basis of
morphological and biochemical characteristics by using direct microscopic
examination and in vitro culture. Although they are economic, these
procedures suffer from the drawbacks of being either slow or non specific
showing false negative results (Howell et al., 1999)[33].
Also the application of chemotherapy has contributed to the occasional
modification and alteration of the morphological characteristics of
dermatophytes cultures and complicating laboratory identification procedures
based on phenotypic features (Faggi et al., 2001)[22].
Using molecular methods for differentiation between the genotypic
characteristics of the species of dermatophytes are more specific, precise,
rapid and are less likely to be affected by external influences such as
temperature variations and chemotherapy. These molecular methods, such as
restriction fragment length polymorphism analysis of mitochondrial DNA (Kano
et al., 2000)[36], sequencing of the
internal transcribed spacer (ITS) region of the ribosomal DNA, sequencing of
protein- encoding genes, and polymerase chain reaction (PCR); random
amplification of polymorphic DNA [RAPD], arbitrarily primed PCR [AP-PCR] (Elisabetta
et al., 2001)[18], and PCR fingerprinting,
have brought important progress in distinguishing between species and
strains. However, most of these techniques require additional manipulation
such restriction endonuclease digestion, hybridization or sequencing after
amplification, so they are complex, laborious, relatively time-consuming,
and not easily employable for routine identification of dermatophytes (Graser
et al., 1999)[29]. In contrast, AP-PCR
technology is simple, rapid and in the absence of specific nucleotide
sequence information for many dermatophyte species, AP-PCR is able to
generate species- specific or strain specific DNA polymorphism on the basis
of characteristic band patterns detected by agarose gel electrophoresis (Faggi
et al., 2001[22]& Baeza and Giannini, 2004[7]).
Dermatophytes are eukaryotic and have machinery for protein and nucleic acid
synthesis similar to that of higher animals. It is, therefore, very
difficult to find out compounds that selectively inhibit fungal metabolism
without exhibiting any toxicity to humans. Also there is evidence that
dermatophytes have acquired resistance to certain antimycotic drugs. So,
with the increasing variety of drugs available for the treatment of
dermatophytoses and with the lack of effective and safe antifungal , the
need for a reference method for testing of antifungal susceptibilities of
dermatophytes has become apparent. Such standard method is not yet
available. Establishment of a reference susceptibility testing method may
allow the clinician to select the appropriate therapy for the treatment of
infections caused by dermatophytes (Jessup et al., 2000 [35]
and Augustine et al., 2005[5]). Aim of
the Study
The current study was conducted to compare the rapid diagnostic molecular
technique AP-PCR with the conventional culture method for identification of
the dermatophyte fungal infections of hair, nail and skin. In addition, to
determine the antifungal susceptibility pattern of different dermatophyte
isolates to Terbinafine, Griseofulvin, Itraconazole and Ketoconazole as the
routinely used antifungal agents. Patients and Methods
The present study was conducted on 115 patients attending the Dermatology
Outpatient Clinic of Ain Shams University Hospitals. Their age ranged from 3
to 50 years (mean 19.8 ± 12.5 SD). They were 59 females and 56 males. The
patients were clinically diagnosed as having tinea capitis (47), tinea
corporis (29), tinea pedis (23), onychomycosis (9), tinea cruris (5) and
tinea manuum (2). All patients were subjected to full history taking
including age, sex and antifungal treatment. Specimen collection and
processing: Clinical examination was done to differentiate between
different types of ring worm infection. Specimens were collected from
all patients after disinfection with 70% alcohol and were kept in dry
sterile containers. The collected specimens were:
- Hair: suspiciously infected hairs were plucked with forceps.
- Cutaneous skin scales: scrapings were taken from the definite edge of
the lesions with sterile scalpel.
- Nail: nail eclipses, scrapings or subangular curette. The specimens
were then subjected to the following (Milne, 2001)[50]:
1. Direct microscopic examination using 10-20% KOH solution with methylene
blue.
2. Culture on:
a) Sabouraud's dextrose agar medium with chloramphenicol (Oxoid, U.K.).
b) Dermasel agar medium with chloramphenicol and cycloheximide (Oxoid,
U.K.).
3. Subculture on:
c) Potato dextrose agar ( PDA )with chloramphenicol (Oxoid,
U.K.).
d) Oatmeal cereal agar for antifungal susceptibility
(Sigma Chemicals Co., St., Louis, Mo., U.S.A.).
e) Potato agar
slants for storage of dermatophyte isolates at -80oC until time of
use.
Identification of the fungal isolates was done through
1. Macroscopic examination of colonies on different media
2. Microscopic examination of cultures.
3. Physiological tests including urease and pigment production tests.
4. Arbitrarily primed polymerase chain reaction (AP-PCR).
Antifungal susceptibility testing of dermatophyte isolates:It was
done according to the standard microdilution method for filamentous fungi M38-A of the
National Committee for Clinical Laboratory Standards (NCCLS, 2002) against
four antifungal agents; Terbinafine, Griseofulvin, Itraconazole and
Ketoconazole with some modifications according to Favre and colleagues
(2003)[23]. I. Arbitrarily Primed
Polymerase Chain Reaction (AP-PCR): Principle: Arbitrarily primed PCR
technique relies on arbitrarily designed sequences of short primers (usually
10 nucleotides long) that anneal specifically to DNA templates in a target
organism. If these primers anneal to target DNA sequences, the intervening
segments that are proximal enough to these annealing sites will be amplified
and will generate products of variable molecular weights. Such products can
be resolved by agarose gel electrophoresis, which will display a band
pattern for each strain. The primer sequences are determined empirically,
since the target DNA are usually not known and the entire genome of the
organism serves as the target for the strain comparison and differentiation
(Liu et al., 2000a [43]& Rilley, 2004[60]).
Procedure: The procedure was done according to Liu et al. (2000b)[44].
a-DNA extraction and precipitation: Fungal isolates were subcultured in
100 ml of Sabouraud's broth (Oxoid, UK) and incubated with shaking for up
to 7 days at 25 °C. Hyphal growth was harvested by filtration and washed
twice with 100ml of sterile saline. Strains, which could not be processed
immediately, were frozen at -80°C prior to extraction. Liquid nitrogen was
added to 2-3g of frozen hyphae and the cells were ground finely.
Approximately 200mg of frozen ground mycelium was placed in a 1.5 ml
microcentrifuge tube. Fungal DNA was extracted from fungal cell suspension
by using the Puregene DNA purification kit (supplied by Gentra system,
U.S.A.). b-cDNA amplification: The primer pair used for Dermatophyte
DNA amplification were OPAA 17 (5'-GAGCCCGACT-3') were prepared by ABI
Applied Biosystem 394 DNA, RNA synthesizer, USA. Six microliters of cDNA
was added with 1.0μl of the primers sense and antisense (50pM/µl) to 50 l
reaction mixture. The reaction mix included 5µl of 1 PCR buffer (50mM KCl,
10mM Tris-HCl, pH 8.3), 2.5mM MgCl2, 0.25μM dATP, 0.25 μM dCTP, 0.25 μM dGTP,
0.75 μM dUTP, 0.125 U of UNG, and 2U of Taq polymerase (0.4µl) (Promega,
USA) and Sterile nuclease free water (38.6µl). After initial denaturation
at 95 C for 2 min, the GenAmp 9700 thermocycler was programmed for 30 cycles
of amplification and a final extension period of 5 min at 72oC. Each cycle
consists of:
* denaturation at 94°C for 30s,
* annealing at <40°C for
30s, and
* extension at 72°C for 30s. The amplified product were then
stored at -20°C . Negative control of sterile nuclease-free water and
positive control of Trichophyton (T). mentagrophytes var erinacei identified
by conventional methods, were included in the procedure. c-Detection and
interpretation of the amplified products: A100-bp DNA ladder (Pharmacia
Biotech, U.S.A.) was used as a molecular size marker. Ten microliters of the
PCR amplicon was mixed with 2μl of gel loading dye (Promega, U.S.A.) and the
mixture was electrophoresed on 1.5% agarose gel in Tris-acetate EDTA buffer
and stained with ethidium bromide (Amersco, U.S.A.). The gel was viewed
under ultraviolet light and photographs were taken. The high intensity bands
produced by AP-PCR for each isolate and the positive control were compared
with the bands of the DNA ladder (figure 1).
 |
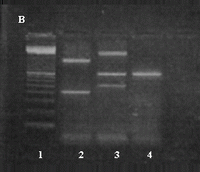 |
Fig 1:
Agarose gel electrophoresis of PCR products of the amplified
Dermatophytes DNA
Lane 1A&B: 100-bp DNA ladder
Lane 2A: T. verrucosum; 500bp
Lane 3A: M. canis; 700, 1200, 3000bp
Lane 4A: M. canis; 700, 1200, 3000bp
Lane 5A: T. rubrum; 400, 600, 2500, 3400bp
Lane 6A: negative control
Lane 2B: T. mentagrophytes var, erinacei; 400, 1500bp
Lane 3B: T. ajelloi; 500, 800, 1900bp
Lane3B: E. floccosum; 800bp |
|
The identification and
differentiation of the 53 isolates were done by comparing the molecular size
of the DNA bands (bp) of the amplified arbitrarily primed PCR products with
that performed by Liu et al. (2000b)[44]
and with the culture results. Table (1) shows the examination of the DNA
products from dermatophyte fungi by AP-PCR.
|
Dermatophyte species and varieties |
Number tested |
DNA products (bp) obtained with random primer OPAA17 |
|
T. rubrum |
18 |
400, 600, 2500, 3400 |
|
T. violaceum |
9 |
400, 1100, 2600, 3500 |
|
T. mentagrophytes var. mentagrophytes |
6 |
2800, 3500 |
|
T. schoenleinii |
4 |
900, 3400 |
|
M. audouinii |
3 |
1300, 2000, 3000 |
|
M. canis |
3 |
700, 1200, 3000 |
|
M. ferrugineum |
2 |
1300, 3000 |
|
T. tonsurans |
2 |
900, 2800, 3400 |
|
E. floccosum |
2 |
800 |
|
T. mentagrophytes var. interdigitale |
1 |
1300, 2800, 3500 |
|
T. mentagrophytes var. erinacei |
1 |
400, 1500 |
|
T. ajelloi |
1 |
500, 800, 1900 |
|
T. verrucosum |
1 |
500 |
Table (1): Examination of DNA products from dermatophyte fungi by AP-PCR obtained by OPAA17 primer (Liu et al., 2000b)[44]
II. Antifungal Susceptibility Testing of Dermatophyte
isolates: (NCCLS, 2002): A standard method for antifungal susceptibility
testing of Dermatophytes is not yet available, mainly the NCCLS (M38-A)
standard method for conidium forming filamentous fungi was followed with
some modifications recommended by Favre et al., (2003)[23]
such as changing temperature and incubation time. a) Antifungal agents:
Terbinafine (Novartis Pharma, Basel, Switzerland), Griseofulvin (Pharco
Pharmaceutical- Alexandria), Itraconazole and Ketoconazole (Janssen- Cilag
Beerse, Belgium) were used in this study. Stock solutions of Terbinafine,
Itraconazole and Ketoconazole were prepared by dissolving 16µg of each
antifungal in 10ml of 100% dimethyl sulfoxide (DMSO) (Sigma chemicals Co.,
St. Louis, Mo., U.S.A.) in separate tubes to get a concentration of
1600µg/ml. For terbinafine 5% Tween 80 was added to the 100% DSMO. While a
stock solution of Griseofulvin was prepared by dissolving 32µg in 10ml of
100% DMSO to get a concentration of 3200µg/ml. Stock solutions were kept
frozen in 1ml aliquots at -70oC. A working solution of each antibiotic was
prepared by diluting 100 µl of the stock solution in 900 µl of RPMI-1640
medium containing L-glutamine and 0.165M morpholine propane sulfonic acid
(MOPS) without bicarbonate (GIBCO BRL, Life Technologies, Paisley, Scotland)
to get a concentration of 32µg/ml for Terbinafine, Itraconazole and
Ketoconazole and 64 µg/ml for Griseofulvin. b) Quality Control Strains:
Candida parapsilosis ATCC 22019 (The American Type Culture Collection (ATCC)
(Rockville, Md) was used as quality control strain to test for the used
antifungal drugs. According to NCCLS M27-A standard method (2000) for
antifungal susceptibility of yeast, the reference MIC range for C.
parapsilosis is 0.06-0.5 µg/ml for both itraconazole and ketoconazole after
48h incubation (Jessup et al., 2000)[35].
Reference strain was grown in 10ml brain heart infusion broth (Difco) at
35oC overnight. The suspension was diluted two folds with brain heart
infusion broth containing 20% glycerol (Sigma), dispensed in screw-capped
tubes, sealed and stored at -70oC. The reference strain was tested with
every batch of antifungal susceptibility of the isolated species (Gupta and
Kohli, 2003)[30]. c) Preparation of the
microdilution plates: o Serial two fold dilution of the antifungal agents
were prepared with RPMI 1640 medium. The final concentrations of the
antifungal agents ranged from 64 to 0.125 for Griseofulvin and from 32 to
0.06µg/ml for Itraconazole, Terbinafine and Ketoconazole. Sterility control
(negative control) and growth control (positive control) were included in
each plate. With each batch of antifungal susceptibility, antifungal control
using C. parapsilosis ATCC 22019 reference strain was inoculated to test for
the validity of the four antifungal agents according to the NCCLS M27-A
standard method (NCCLS, 1997). o Uninoculated microtitration plates
containing antifungal dilutions were kept covered for approximately 6 months
at -70°C. d) Preparation of the Dermatophyte inoculum: Dermatophyte
isolates were grown on oatmeal cereal agar slants for 7 days at 28°C; the
best medium to support conidial growth (Jessup et al., 2000)[35].
Sterile normal saline (0.85%) was added to the slant culture and was gently
swabbed with a cotton tip applicator to dislodge the conidia from the hyphal
mat. The suspension was adjusted to 5 mL with sterile normal saline. The
cell density was adjusted to give final inoculum concentration of 104CFU/ml.
The suspension was counted on a hemocytometer and was diluted in RPMI 1640
to the desired concentration. 100µ1 of the organism suspension was
transferred into all wells of the microdilution plates except for the
negative control wells. Plates were incubated aerobically at 30°C, except
for E. floccosum and M. canis at 35°C. All were incubated for 4-10 days
according to the growth in the control wells. e) Reading and
interpretation of the panel: The minimal inhibitory concentration (MIC)
endpoints were determined according to NCCLS M38-A standards as the point at
which no visual turbidity where the organism was inhibited 80% when compared
to the growth control. For the quality control Candida parapsilosis the MIC
endpoint was determined as
≥80%
inhibition of the positive growth control for Itraconazole and Ketoconazole
(NCCLS M27-A). Results
This study was carried out on 115 patients with
ringworm infection. Their age ranged from 3-50 years. Forty seven patients
were below age of ten years (41%), eleven were in the second decade (9.4%),
thirty one patients were in the third (27%), twenty three patients were in
the fourth (20%) and three patients were in the fifth decade (2.6%). The
patients under study were 59 (51.3%) females and 56 (48.7%) males. Tinea
capitis infection 47 (41%) was only prevalent below the age of ten years and
double in females 31 (27%) than males 16 (13.9%). Also tinea pedis was
higher in males 13 (11.3%) than females 10 (8.7%). Table (2) shows the
distribution of the Dermatophyte positive cultures among patients with
ringworm infection. Tinea capitis represented 47 (41%) of cases followed by
tinea corporis 29 (25.2%) and tinea pedis 23 (20%). Twenty three (43%) of
dermatophyte isolates were separated from tinea capitis patients, followed
by 14 (26.4%) and 12 (22.6%) from tinea corporis and tinea pedis
respectively.
|
Diagnosis |
Clinical Cases |
Positive Cases by culture |
|
No. |
% |
No. |
% |
|
T. capitis |
47 |
41.00% |
23 |
43.40% |
|
T. corporis |
29 |
25.20% |
14 |
26.40% |
|
T. pedis |
23 |
20.00% |
12 |
22.60% |
|
Onychomycosis |
9 |
7.80% |
2 |
3.80% |
|
T. cruris |
5 |
4.30% |
1 |
1.90% |
|
T.mannum |
2 |
1.70% |
1 |
1.90% |
|
Total |
115 |
100.00% |
53 |
100.00% |
Table (2): Distribution of Dermatophyte Positive Cultures Among
Patients with Skin fungal Infection
In spite that there was no statistically significant
association between the isolation of dermatophytes from the infected sites
and the sex of the patients (P>0.5), dermatophytes isolated from tinea
capitis patients were higher in females 15 (28.3%) than males 8 (15.1%).
Males show higher prevalence of tinea pedis than females. Out of the 115
specimens of the patients under study, 53 (46.1%) yielded dermatophyte growth
on culture and 21 (18%) specimens grew nondermatophytes; nine Candida, eight
Aspergillus spp. (five A. fumigatus, three A. niger) and four cases showed
Acremonium. Three (2.61%) of the patients showed both Dermatophytes and non
dermatophytes growth. The later group grew mainly on Sabouraud's dextrose
agar (SDA). In the present study there was a highly significant
association (P<0.001) between the results of direct microscopic examination
by KOH and dermatophyte culture. Out of the 53 positive cases by culture,
there were 38 positive cases (33%) by direct KOH examination and the
remaining 15 cases (13%) were negative and out of 62 negative cases by
culture, there were 16 (14%) cases positive by direct KOH examination. Out
of those 16 dermatophyte culture negative specimens and KOH positive; five
cases were on antimycotic therapy (two tinea cruris and three tinea
capitis), five cases grew Aspergillus (three obtained from tinea capitis and
two from onychomycosis), five cases Candida (two were onychomycosis, one
tinea pedis, one tinea capitis and one tinea cruris) and one case of tinea
capitis showed mixed growth of Aspergillus and Candida. The KOH method had a
sensitivity of 88% and specificity of 74%. By direct KOH microscopic
examination no definite identification was reached. However, hyphae
,arthrospores and chlamydospores were seen in some preparations. Acremonium
species gave the fronded appearance and Aspergillus branched dichotomously
at acute angles. Out of the three cases that yielded growth of both
dermatophytes and rapidly growing non Dermatophyte fungi, one was
onychomycosis which showed the fruiting bodies of Aspergillus together with
the hyphae of dermatophytes on KOH examination. The second two cases were
tinea corporis which showed budding yeast cells of Candida species on direct
microscopy together with the hyphae of dermatophytes. Table (3) shows the
dermatophyte species isolated from each type of fungal infection. The most
common type was T. rubrum 17 (32.1%) which showed a highly significant
association with almost all types of dermatophytosis except tinea capitis
(P<0.001). There were a highly significant association between T.
mentagrophytes 11 (20.8%), T. violaceum 10 (18.9%) with tinea capitis,
corporis and pedis patients (P<0.001).
|
Species of Dermatophytes |
T. capitis |
T. corporis |
T. pedis |
T. unguium No (%) |
T. cruris
No (%) |
T. mannum No (%) |
Total No (%) |
|
No (%) |
No (%) |
No (%) |
|
T. rubrum |
0 |
5 (35.7%) |
8 (66.7%) |
2 (100.0%) |
1(100.0%) |
1 (100.0%) |
17 (32.1%) |
|
T. mentagrophytes |
7 (30.4%) |
2 (14.3%) |
2 (16.7%) |
0 |
0 |
0 |
11 (20.8%) |
|
T. violaceum |
6 (26.1%) |
3 (21.4%) |
1 (8.3%) |
0 |
0 |
0 |
10 (18.9%) |
|
T. verrucosum |
1 (4.3%) |
0 |
0 |
0 |
0 |
0 |
1 (1.9%) |
|
T. schoenleinii |
2 (8.8%) |
0 |
0 |
0 |
0 |
0 |
2 (3.8%) |
|
T. soudanense |
1 (4.3%) |
0 |
0 |
0 |
0 |
0 |
1 (1.9%) |
|
T. tonsurans |
0 |
1 (7.1%) |
0 |
0 |
0 |
0 |
1 (1.9%) |
|
M. canis |
2 (8.8%) |
1 (7.1%) |
0 |
0 |
0 |
0 |
3 (5.6%) |
|
M. ferrugineum |
1 (4.3%) |
1 (7.1%) |
0 |
0 |
0 |
0 |
2 (3.8%) |
|
M. audouinii |
3 (13.0%) |
0 |
0 |
0 |
0 |
0 |
3 (5.6%) |
|
E. floccosum |
0 |
1 (7.1%) |
1 (8.3%) |
0 |
0 |
0 |
2 (3.8%) |
|
Total No. |
23
(100%) |
14
(100%) |
12
(100%) |
2 (100%) |
1
(100%) |
1 (100%) |
53
(100%) |
Table (3): Dermatophytes Species
Isolated From different types of fungal infections by culture Table (4) shows the
results of the phenotypic and genotypic identification of dermatophyte
isolates. Out of the 115 specimens the highest rate of dermatophyte
isolation on primary culture media was on Dermasel agar (46%) after
incubation at 28oC, followed by incubation at 37oC (12%) then SDA (7%) at
28oC. T. mentagrophytes and M. canis were the fastest to grow on Dermasel
media (primary culture media) (6 days) at 28°C while T. violaceum was
the slowest to grow (23days). All dermatophytes grew better on 28°C
with exception of T. violaceum and T. verrucosum which grew faster at 37°C .
Subculture of dermatophyte isolates on PDA showed that the fastest growth
was for T. mentagrophytes and M. canis (3 days) and the slowest were for
T. violaceum, T. schoenleinii and T. soudanense (10 days). Comparison
between the identification of dermatophyte isolates from culture by the
ordinary phenotypic methods (morphology and physiological tests) and the
genotypic method by the AP-PCR was done in this study. The genotypic
identification was in agreement with the conventional phenotypic methods in
46 (86.8%) out of 53 Dermatophyte isolates while the disagreement was in 7
cases (13.2%). Out of the eleven isolates of T. mentagrophytes only two were
diagnosed phenotypically to the strain level and confirmed genotypically,
var. mentagrophytes and var. erinacei. By the genotypic AP-PCR five were
found to be var. mentagrophytes and one var. interdigitale. The last three
isolates, two of them were diagnosed by PCR as T. rubrum, and one as T.
tonsurans. In addition two isolates of T. violaceum were diagnosed by PCR as
T. schoenleinii. One T. rubrum isolate was genotypically diagnosed as T.
ajelloi. The last T. soudanense isolate was genetically diagnosed as T.
violaceum.
|
No. |
Primary Culture media |
PDA Sub-culture |
Culture Results |
PCR Results |
|
SDA |
SDA |
DS |
DS |
|
28°C |
37°C |
28°C |
37°C |
|
2 |
-ve |
-ve |
7d |
-ve |
3d |
T. mentagrophytes |
T.ment v. mentagrophytes |
|
3 |
-ve |
-ve |
9d |
-ve |
6d |
T. mentagrophytes* |
T. rubrum |
|
6 |
-ve |
-ve |
16d |
-ve |
9d |
T. rubrum |
T. rubrum |
|
7 |
8d |
-ve |
7d |
-ve |
3d |
T.ment v. mentagrophytes |
T.ment v. mentagrophytes |
|
11 |
-ve |
-ve |
20d |
17d |
7d |
T. violaceum* |
T. schoenleinii |
|
12 |
-ve |
-ve |
7d |
-ve |
5d |
T. mentagrophytes |
T.ment v. mentagrophytes |
|
15 |
15d |
16d |
14d |
16d |
7d |
T. rubrum |
T. rubrum |
|
21 |
Candida |
Candida |
15d |
-ve |
8d |
T. violaceum |
T. violaceum |
|
22 |
Candida |
-ve |
11d |
-ve |
7d |
T. rubrum |
T. rubrum |
|
28 |
-ve |
-ve |
7d |
8d |
5d |
M. audouinii |
M. audouinii |
|
31 |
-ve |
-ve |
6d |
-ve |
6d |
T. mentagrophytes |
T.ment v. mentagrophytes |
|
32 |
-ve |
-ve |
9d |
13d |
6d |
E. floccosum |
E. floccosum |
|
34 |
-ve |
-ve |
6d |
-ve |
3d |
M. canis |
M. canis |
|
39 |
-ve |
-ve |
23d |
-ve |
10d |
T. violaceum |
T. violaceum |
|
40 |
11d |
-ve |
19d |
-ve |
9d |
T. violaceum |
T. violaceum |
|
41 |
-ve |
-ve |
15d |
-ve |
10d |
T. violaceum |
T. violaceum |
|
42 |
-ve |
-ve |
10 d |
-ve |
8d |
T. rubrum |
T. rubrum |
|
45 |
-ve |
-ve |
12d |
16d |
8d |
T. mentagrophytes* |
T. rubrum |
|
46 |
-ve |
-ve |
12d |
15d |
9d |
T. rubrum |
T. rubrum |
|
47 |
-ve |
-ve |
10d |
-ve |
7d |
T. rubrum |
T. rubrum |
|
52 |
-ve |
-ve |
17d |
-ve |
10d |
T. violaceum |
T. violaceum |
|
53 |
12d |
-ve |
10d |
-ve |
7d |
T. tonsurans |
T. tonsurans |
|
55 |
-ve |
-ve |
15d |
-ve |
8d |
T. violaceum* |
T. schoenleinii |
|
58 |
14d |
-ve |
10d |
-ve |
5d |
E. floccosum |
E. floccosum |
|
61 |
-ve |
-ve |
7d |
-ve |
4d |
T. mentagrophytes |
T.ment. v. interdigitale |
|
65 |
-ve |
-ve |
15d |
-ve |
8d |
T. violaceum |
T. violaceoum |
|
68 |
-ve |
-ve |
10d |
10d |
6d |
T. rubrum |
T. rubrum |
|
71 |
-ve |
-ve |
6d |
7d |
4d |
T. mentagrophytes* |
T. tonsurans |
|
72 |
-ve |
-ve |
6d |
-ve |
4d |
M. canis |
M. canis |
|
73 |
-ve |
-ve |
17d |
-ve |
10d |
T. schoenleinii |
T. schoenleinii |
|
75 |
-ve |
-ve |
10d |
-ve |
7d |
T. rubrum |
T. rubrum |
|
77 |
10d |
-ve |
8d |
-ve |
5d |
M. ferrugineum |
M. ferrugineum |
|
79 |
-ve |
-ve |
11d |
-ve |
7d |
M. audouinii |
M. audouinii |
|
82 |
-ve |
-ve |
20d |
-ve |
10d |
T. schoenleinii |
T. schoenleinii |
|
84 |
-ve |
-ve |
14d |
13d |
9d |
T. rubrum |
T. rubrum |
|
86 |
15d |
-ve |
10d |
12d |
7d |
T. rubrum |
T. rubrum |
|
87 |
-ve |
-ve |
17d |
-ve |
9d |
M. ferrugineum |
M. ferrugineum |
|
88 |
-ve |
-ve |
9d |
-ve |
5d |
T. mentagrophytes |
T.ment v. mentagrophytes |
|
89 |
-ve |
-ve |
10d |
-ve |
5d |
T. rubrum |
T. rubrum |
|
92 |
10d |
-ve |
7d |
-ve |
4d |
M. canis |
M. canis |
|
93 |
-ve |
-ve |
22d |
-ve |
10d |
T. soudanense* |
T. violaceum |
|
95 |
-ve |
9d |
7d |
-ve |
5d |
T.ment. v. erinacei |
T.ment var. erinacei |
|
98 |
-ve |
-ve |
20d |
22d |
10d |
T. violaceum |
T. violaceum |
|
101 |
-ve |
-ve |
10d |
12d |
8d |
T. rubrum |
T. rubrum |
|
103 |
-ve |
-ve |
10d |
-ve |
7d |
T. rubrum |
T. rubrum |
|
105 |
-ve |
-ve |
19d |
17d |
8d |
T. verrucosum |
T. verrucosum |
|
106 |
-ve |
-ve |
19d |
-ve |
9d |
T. violaceum |
T. violaceum |
|
107 |
Aspergillus |
Aspergillus |
11d |
-ve |
6d |
T. rubrum |
T. rubrum |
|
108 |
-ve |
-ve |
10d |
-ve |
6d |
M. audouinii |
M. audouinii |
|
109 |
-ve |
-ve |
6d |
8d |
4d |
T. mentagrophytes |
T.ment v. mentagrophytes |
|
111 |
-ve |
-ve |
12d |
-ve |
7d |
T. rubrum* |
T. ajelloi |
|
114 |
-ve |
-ve |
10d |
-ve |
9d |
T. rubrum |
T. rubrum |
|
115 |
-ve |
-ve |
10d |
-ve |
7d |
T. rubrum |
T. rubrum |
Table (4): Results of Phenotypic and Genotypic Identification
of Dermatophyte isolates Table (5) shows the pattern of antifungal
susceptibility of the dermatophyte isolates and the range of their MIC
endpoints by broth microdilution method (NCCLS M38-A with modifications)
towards the four antifungal drugs. The MIC ranges (mean) were as follows:
for terbinafine from 0.06 to 0.5 (0.121) µg/ml, itraconazole from 0.06 to 4
(0.62) µg/ml, ketoconazole ranged from 0.06 to 4 (0.857) µg/ml and griseofulvin from 0.5 to 8 (2.151)
µg/ml. Terbinafine was the most powerful
antimycotic. T. highest MIC values for the four antifungal agents.
|
Rubrum had the Dermatophytes Species |
MIC endpoints |
|
Terbinafine |
Itraconazole |
Griseofulvin |
Ketoconazole |
|
T. mentagrophytes |
0.06 - 0.25 |
0.125 -2 |
1 - 4 |
0.25 -2 |
|
T.ment,var erinacei |
0.06 - 0.125 |
0.25 – 0.5 |
0.5 - 2 |
0.06 - 0.25 |
|
T. rubrum |
0.06 - 0.5 |
0.125 - 4 |
1 - 8 |
0.06 – 4 |
|
T. violaceum |
0.06 - 0.125 |
0.06 – 0.5 |
0.5 - 2 |
0.06 - 0.5 |
|
T. verrucosum |
0.06 |
0.125 |
2 |
1 |
|
T. schoenleinii |
0.06 |
0.125 - 0.5 |
0.5 - 1 |
0.125 |
|
T. soudanense |
0.06 |
0.5 |
2 |
1 |
|
T. tonsurans |
0.5 |
2 |
1 |
4 |
|
M. canis |
0.06 -0.5 |
0.125 – 2. |
2 - 4 |
0.25 – 1 |
|
M. ferrugineum |
0.06 - 0.125 |
0.06 - 0.125 |
0.5 |
0.125 |
|
M. audouinii |
0.06 - 0.125 |
0.06 - 0.125 |
1 - 2 |
0.125 - 2 |
|
E. floccosum |
0.06 |
0.125 |
4 |
2 |
|
Range |
0.06 -0.5 |
0.06 – 4 |
0.5 - 8 |
0.06 – 4 |
|
Mean |
0.121 |
0.620 |
2.151 |
0.857 |
|
SD |
0.122 |
0.973 |
1.637 |
0.991 |
Table
(5): Antifungal Susceptibility Pattern of Dermatophyte isolates by NCCLS
M38-A method Discussion
With the increasing incidence
and mortality of fungal infection, the requirement for strict diagnostic
approaches became a very urgent issue. In addition, the traditional
detective techniques, such as culture, give poor diagnostic approaches,
accordingly, the molecular tools of classification and identification of
pathogenic fungi such as dermatophytes considered of great help (Li et al.,
2004)[41]. As fungal infection of the
skin is the most common infectious dermatologic condition throughout the
world (Gupta et al., 2003)[31] and because
of the difference in distribution of these types of infections from country
to country (Chan and Friedlander, 2004)[12],
our study was done to detect Dermatophytes distribution in Egyptian
patients. In this study, tinea capitis accounted for 41% of all cases,
followed by tinea corporis (25.2%), tinea pedis (20%), onychomycosis (7.8%),
tinea cruris (4.3%) and tinea manuum (1.7%). This copes with results done by
Omar, (2000)[58] who emphasized that tinea
capitis is considered as the major type of fungal infection in Egypt. In
Libya, tinea corporis accounted for 45.9% of cases followed by tinea pedis
(8.1%) and tinea manuum (2.6%) as discussed by Ellabib et al., (2002)[19].
In Yemen, tinea corporis accounts for the majority of cases followed by
tinea capitis as mentioned by Mahmoud, (2002)[45].
The endemic nature of scalp infection in the developing countries is
perpetuated by lack of knowledge about prevalence, carrier state, and the
absence of control measures (Moubasher et al., 2000)[51].
In the developed countries, tinea pedis is the major type of fungal
infection. In Japan Takahashi and Nishimura, (2002)[66]
revealed that tinea pedis accounts for 64.2%, followed by tinea unguium
(14.6%), tinea corporis (11.9%), tinea cruris (5.4%), tinea manuum (3.6%)
and tinea capitis (0.2%). This was assured by results of Seebacher, (2003)[61],
who found increase in the incidence of tinea pedis in Central and North
Europe. Aste et al., (2003)[4] reported that
tinea pedis accounts for 23.4% in Italy. Wearing shoes for long time as in
athletes and excessive use of water in gymnasiums will result in maceration
of toe clefts predisposing to tinea pedis infection (Tietz, 2003[68]).
In the present study, tinea capitis was found only in children below the age
of ten years (41%). Our results were in accordance with the studies done by
Omar (2000)[58] in Alexandria (54%), Fuller
et al., (2003)[28] in south-east London and
by Mounkassa, et al., (2004)[52] in France
(70%). Aste, et al., (2003)[4]; Gupta, et
al., (2003)[31]; Hubert and Callen, (2003)[34];
Shibaki and Shibaki, (2003)[62] and
Sigurgeirsson and Steingrimsson, (2004)[63]
all agreed that above the age of ten years tinea infections are other types
than tinea capitis, a fact that was explained by the started sebum
production about this age, with its antifungal properties. In this study,
tinea capitis was double in female patients than males (27% and 13.9%
respectively) although there were no statistically significant correlation
with sex ( P>0.05). This was in accordance with Omar (2000), who found that
tinea capitis was more common in school girl children. This may be due to
sharing combs and sharing facilities in hair dresser. This result is
different from that done in Kenya, by Ayaya et al. (2001)[6],
who found that tinea capitis was higher in boys (60.9%) than girls (39.1%),
this may be due to some bad habits as sharing caps and combs, and it cannot
be correlated with specific sex. On the other hand, our results showed that
the rate of tinea pedis, was higher in males than females (11.3% and 8.7%
respectively). This goes with the results done by Sofia et al., (2001)[64]
and Cheng and Chong, (2002)[13]. Aste, et
al., (2003)[4] reported that adult males
probably have about 20% chance of developing tinea pedis, while among women
only 5% are likely to become clinically infected, because it occurs more
common in athletes sharing washing facilities and using common swimming
baths. In our country males use sport tight shoes more frequently than
females. Males also wear closed shoes for long time in comparison with
females. Dermatophyte and nondermatophyte species were isolated in this
study from 46.1% and 18% of the cases of fungal infection respectively.
These results are nearly similar to the results reported by Al-Sogair et al.
(1991)[1] in Saudi Arabia, who isolated both
dermatophytes and nondermatophytes in 57.2% of cases. Candida and
Aspergillus were the main nondermatophytes isolated in their study as in our
study. Infection caused by yeast and moulds were involved in our study
because no marked differences in their clinical picture from that of
dermatophyte infection could be observed. This reveals the role of
nondermatophytes species in infection of the skin and its appendages . In
the present study, out of the positive specimens by culture, 71.7% were
positive by the direct KOH microscopic examination; two of them showed mixed
dermatophytes and nondermatophytes infection. The KOH direct method had a
sensitivity of 88% and specificity of 74%. This was in agreement with
Escobar and Carmona-Fonseca (2003)[20] and
Arca et al. (2004)[3], who found that direct
microscopy was positive in 92% and 77% respectively from all culture
positive cases. In our study direct microscopic examination show false
negative results in 13% of cases. This is in accordance with Liu et al.
(2000b)[44], who stated that this method is
insensitive, showing false negative results up to 15%. While Tampieri,
(2004)[67], mentioned that direct
microscopic examination shows false negative results up to (50%). This may
be because direct microscopic examination appears to be greatly influenced
by the meticulous preparation of specimens, experience of the observer and
rate of contamination. On the other hand there were 16 (14%) positive
specimens by direct KOH examination that showed negative results by culture
This may be because 5/16 cases were on antimycotic treatment so gave no
growth on culture. The remaining 11/16 cases showed either true mixed
infection or contamination by the rapidly growing fungi (Aspergillus,
Candida and Acremonium) than dermatophytes that covered the entire medium
giving no chance for the slowly growing dermatophytes to appear. In our
study, Dermatophytes were seen on microscopic examination as hyphae,
arthrospores and chlamydospores. Acremonium species gave the fronded
appearance and Aspergillus branched dichotomously at acute angles. However
no definite identification was reached. In agreement with Liu et al.,
(1997)[42], Elweski (1995)[17]
and Escobar and Carmona-Fonseca, (2003)[20] it seems to be
difficult to rely on results of direct microscopy with KOH to establish the
diagnosis of fungal infection as it could not detect the characteristic
morphology of the three genera and it lacks sufficient sensitivity but, it
is highly efficient as screening technique before therapy is initiated
because of the expense, duration and potential adverse effects of the
treatment (Tampieri, 2004 [67]& Hainer,2003[32]).
In this study Trichophyton rubrum was the most frequently isolated organism
(32.1%) mainly from tinea pedis and tinea corporis. These results were in
agreement with those detected in France (35.5%) (Lacroix et al., 2002)[39].
While higher isolation rates of T. rubrum were reported in Australia (69.5%)
(Coloe and Baird, 1999)[14], Japan (79.4%)
(Shibaki and Shibak, 2003) and Slovakia (81.6%) as reported by Buchvald and
Simaljakova, (2002)[10]. On the other hand,
in Libya, isolation rate of the organism was lower (13%) as mentioned by
Ellabib et al., (2002)[19]. No isolates of
T. rubrum was detected from tinea capitis patients in this study. This was
in accordance with Fisher and Cook (1998)[26]
who reported that T. rubrum is the most common cause of tinea pedis and
rarely causes tinea capitis. The most frequently isolated Dermatophytes
from tinea capitis patients were Trichophyton mentagrophytes (30.4%) and
Trichophyton violaceum (26.1%). In contrast Moubasher et al., (2000)[51],
found that T. violaceum accounts for (62%) of all cases with tinea capitis
while it accounts for (91%) in London as mentioned by Fuller et al., (2003)[28].
In Brazil, Microsporum canis accounts for (71.3%) as discussed by Dias et
al., (2003)[16] and Chan and Friedlander
(2004) and in Germany it was (50%) as mentioned by Lehmann et al., (2004)[40].
In Turkey, T. verrucosum accounts for (43%) in the study done by Metin et
al., (2002)[49]. This can be attributed to
the fact that species of dermatophytes causing tinea capitis vary from
country to country and also change with time, geography,
environment, climate, occupation, ethnic group and life styles ( Nweze and
Okafor 2005)[56]. The highest rate of
Dermatophyte isolation on primary culture media was on Dermasel agar (46%)
after incubation at 28oC, with exception of T. violaceum and T. verrucosum
which grew faster at 37°C . This elevates the importance of dermasel as
selective medium that contains cycloheximide for the isolation of
dermatophytes at 28oC which is the optimum temperature for most of
dermatophytes (Aly, 1994[2] & Hainer, 2003[32]).
Trichophyton mentagrophytes and M. canis were the fastest to grow (6 days)
while T. violaceum was the slowest to grow (23days). Subculture of
Dermatophyte isolates on PDA showed that the fastest growth was for T.
mentagrophytes and M. canis (3 days) and the slowest were for T. violaceum,
T. schoenleinii and T. soudanense (10 days). In-vitro culture is capable of
providing a species-specific determination of dermatophytes on the basis of
morphological and biochemical criteria in 5-15days in >95% of cases.
However, for some unusual and atypical isolates, identification may require
a range of culture media and tests. These tests are costly, time consuming
(3-4 weeks after primary isolation) and demand specialist skills. More
importantly, because these conventional methods depend on measurement of the
phenotypic characteristics of dermatophytes, they can be easily influenced
by outside factors such as temperature variations and chemotherapy that may
affect the interpretation of in-vitro culture results (Liu et al., 2000b)[44].
In the present study, genotypic identification of Dermatophyte isolates by
the AP-PCR using the OPAA-17 primer was in agreement with the phenotypic
methods in 86.8% of the isolates and the disagreement was in 13.2% of them.
Phenotypic identification of T. mentagrophytes to the strain level could
only be done for two isolates form eleven. The disagreement between the two
methods of identification was reported by Kawai, (2003)[37].
These may be explained as the conventional methods of identification require
special skills, or because of atypical culture characteristics due to
treatment. Two isolates of T. mentagrophytes were genotypically diagnosed as
T. rubrum and one as T. tonsurans. This may be explained as the three
strains were similar to T. mentagrophytes in having similar morphology,
moderately slow-grower, and flat, granular, creamy, with reddish brown
reverse. On microscopic examination the genetically diagnosed T. rubrum
strains showed many drop shaped to round microconidia and abundant club
shaped macroconidia. This morphology and genetic characters are matching
with the urease positive Asiatic variant of T. rubrum which is different
from the typical T. rubrum but is minimally genetically distinguished from
it. It is usually separated from tinea corporis. Trichophyton tonsurans
develops several types of colonies with a variety of colors and surface
textures and is urease positive that resembles T. mentagrophytes. They have
branched septate hyphae with terminal swellings, numerous microconidia
forming loose clusters and few short blunt cylindrical macroconidia (Bassiem
and El-Borhamy, 2002[9]& Summerbell, 2003[65]).
Two T. violaceum isolates were diagnosed by PCR as T. schoenleinii and one
T. soudanense was genetically diagnosed as T. violaceum. Fisher and Cook
(1998) recommended that those three species must be differentially diagnosed
from each others by further tests other than the usual KOH and culture
methods. They were all slow growers, developed at first glabrous
cream-colored colonies and as they aged they developed the deep red or
purple color. They all had few macro and microconidia with short segmented
and distorted twisted branching hyphae. Finally one T. rubrum isolate was
genotypically diagnosed as T. ajelloi. T. ajelloi gave purple black pigments,
with smooth thick walled cylindrical macroconidia as T. rubrum. Its presence
may be due to mixed infection with a slow grower organism or due to
contamination of culture as Summerbell (2003)[65]
reported that there is no data to say it is pathogenic. Although most
dermatophytes can be identified after primary isolation (10-15days), a few
may require secondary culture on specialized media (10-15days). With some
atypical, unusual or slow-growing isolates, the identification process may
take even longer time. Therefore, DNA analysis using AP-PCR technique has
clear advantage over conventional techniques for identification of
dermatophytes. PCR made a genetic-based differentiation of dermatophytes
species possible at the species and strain level, more rapid (within one
day), sensitive and more precise and stable. AP-PCR can identify young
culture before development of any characteristic feature of the organism,
dermatophytes with atypical morphology and dead strains (Faggi et al.,
2001)[22]. This is necessary to survey the
current epidemiologic situation and to trace back the pathogenesis of
infection in order to avoid reinfection and thus optimize the therapy. In
the present study, the NCCLS M38-A broth microdilution method was used to
determine the antifungal susceptibility pattern of dermatophytes isolated
from the clinical specimens. The MIC (mean) (µg/ml) endpoints obtained
ranged from 0.06 to 0.5 (0.121) for terbinafine, 0.06 to 4 (0.62) for
itraconazole, 0.5 to 8 (2.151) for griseofulvin and 0.06 to 4 (0.857) for
ketoconazole. Our results were significantly higher than those obtained by
Barros and Hamdan (2005)[8] where MICs were
<0.007-0.015 for terbinafine, 0.062-1.0 for itraconazole, 0.25-2.0 for
griseofulvin and. 0.125-2.0 for ketoconazole. Our results were also higher
than the MIC ranges and means obtained by Pujol et al. 2005[57],
Brilhante et al. (2005)[11] and Esteban et
al. (2005)[21]. These higher MICs than
those of other studies, possibly because of differences in culture medium
used in the other studies or may be due increased use of local and oral
antifungal agents or repeated intake due to patient incompliance or chronic
infection. In this study T. rubrum had the highest MIC values for the four
antifungal agents. This is in accordance with Mukherjee et al. (2003)[53]
who detected T. rubrum strains with primary resistance to terbinafine. Our
study demonstrated that the four antifungal drugs used were active against
dermatophytes, although these results were species dependent which cope with
that obtained by Fernindez et al., (2002)[25].
In the present study terbinafine possessed the highest antifungal activity
against all dermatophytes tested in vitro. This was in accordance with the
results obtained by Fernindez et al. (2001 and 2002)[24,25],
Favre et al. (2003)[23] and Gupta and Kohli
(2003)[30] even though a reference method
for testing dermatophytes still has not been developed. In vivo terbinafine
is the extremely potent systemic drug against dermatophytes and it provides
super long term mycological and clinical efficacy and lower rates of
clinical relapse as mentioned by Darkes et al., (2003)[15].
In conclusion, it seems to be difficult to rely on results of direct
microscopic examination with KOH to establish the diagnosis of different
fungal infections of the skin, as it lacks sufficient sensitivity; however,
it is highly efficient as a screening technique. Culture on the specific
media of dermatophytes will ensure the diagnosis and reach to the species
level and sometimes to the strain level. However it may be time consuming,
costly, as it needs different culture media for proper identification, in
addition it needs special skills and experiences as the morphological
character of some species are atypical. The genotypic differentiation by
AP-PCR provides a rapid and practical tool for identification of
dermatophyte isolates that is independent of morphological and biochemical
characteristics thus, enhances laboratory diagnosis of dermatophytes. AP-PCR
represents a technological advance in the laboratory diagnosis of
dermatophytosis. Further investigation of a larger number of isolates could
shed light upon more details of these kinds of infections. The future
development of procedure for the isolation of DNA from clinical materials
such as skin, hair and nails would further enhance the potential of AP-PCR
for identification of dermatophytes. This study shows that the standard
NCCLS M38-A broth microdilution method with the modifications made in
temperature and incubation period are convenient for antifungal
susceptibility testing of dermatophytes. Among the antifungal tested
terbinafine was the most potent antifungal drug. Further studies are needed
to standardize these optimal growth conditions, ensure reproducibility and
allow the comparison between different antimycotics. MICs need to be
correlated with clinical outcome to develop interpretive breakpoints, which
in turn may clarify the reasons for lack of clinical response and detection
of resistance. This is especially important for immunocompromised patients
and young children. References
1. Al-Sogair,
S.M.; Moawad, M.K. and Al-Humaidan, Y.M. (1991): Fungal infection as a cause
of skin disease in the Eastern providence of Saudi Arabia: prevailing fungi
and pattern of infection. Mycoses 34: 333-37.
2. Aly,
R. (1994): Culture media for growing dermatophytes. J. Am Acad. Dermatol.
31S:107-08.
3. Arca, E., Saracli, M.A., Akar, A.,
Yildiran, S.T. Kurumlu, Z. and Gur, A.R. (2004): Polymerase chain reaction
in the diagnosis of onychomycosis. Eur J Dermatol. Jan-Feb; 14(1):52-5.
4. Aste, N., Pau, M., Aste, N. and Biggio, P. (2003): Tinea pedis observed
in Cagliari, Italy, between 1996 and 2000. Mycosis 46: 38-41.
5. Augustine, S.K.; Bhavsar, S.P. and Kapadnis, B.P. (2005): Production of a
growth dependent metabolite active against dermatophytes by Streptomyces
rochei AK 39. Indian J. Med. Res. 121:164-170.
6.
Ayaya, S.O.; Kamar, K.K.; and Kakai, R. (2001): Aetiology of tinea capitis
in school children. East Afr. Med. J. 78:531-35.
7.
Baeza. L.C. and Giannini, M.J.S.M. (2004): Strain differentiation of
Trichophyton rubrum by random amplification of polymorphic DNA (RAPD).
46:339-41.
8. Barros, M.E.S. and Hamdan, J.S. (2005):
Determination of susceptibility/resistance to antifungal drugs of
Trichophyton mentagrophytes isolates by a macrodilution method. Can. J.
Microbiol./Rev. can. microbiol. 51: 983-87.
9.
Bassiem, H.H. and El-Borhamy, M. (2002): Medical mycology. In Diagnostic
Microbiology (1st ed), p:118-165.
10. Buchvald, J.
and Simaljakova, M. (2002): Epidemiology of dermatomycoses in Slovakia.
Epidemiol Mikrobiol Immunol. Apr; 51(2): 71-3.
11.
Brilhante, R.S.N.; Cordeiro, R.A.; Medrano, D.J.A.; Monteiro, A.J.; Sidrim,
J.J.C. and Rocha, M.F.G.(2005): Antifungal susceptibility and genotypical
pattern of Microsporum canis strains. Can. J. Microbiol./Rev. can. microbiol.
51: 507-10.
12. Chan, Y.C. and Friedlander S.F.
(2004): New treatments for tinea capitis. Current Opin. Infect.
Dis.17:97-103 (Abstract).
13. Cheng, S. and Chong,
L. (2002): A prospective epidemiological study on tinea pedis and
onychomycoses in Hong Kong. Chin. Med. J. 115: 860-65.
14. Coloe, S.V. and Baird, R.W. (1999): Dermatophyte infections in
Melbourne: trends from 1961/64 to 1995/95. Pathology; 31:395-97.
15. Darkes, M.J., Scott, L.J. and Goa, K.L. (2003): Terbinafine: a review of
its use in onychomycosis in adults. Am J Clin Dermatol. 4(1): 39-65.
16. Dias, T., Fernandes, L.; Ode, F.; Soares, A.J.; Passos, X.S.; Costa, M.;
Hashimoto, H.; Souza, L.K.; and Silva, N. and Mdo, R. (2003): Tinea capitis
in children from Goiania, Brazil. Rev. Soc. Bras. Med. Trop. 36:653-55.
17. Elewski, B.S. (1995): Clinical pearl: Diagnosis of onychomycosis. J. Am.
Acad. Dermatol. 32: 500-01.
18. Elisabetta, F.,
Gabriella, P., Enza, C., Chiara, B., Elisa D. and Francesca, M. (2001):
Application of PCR to distinguish common species of dermatophytes. J Clin.
Microbiol. 39: 3382-85.
19. Ellabib, M.S., Khalifa,
Z. and Kavanagh, K. (2002): Dermatophytes and other fungi associated with
skin mycoses in Tripoli, Libya. Mycoses 45: 101-4.
20. Escobar, M.L. and Carmona-Fonseca, J. (2003): Onychomycosis by common
non dermatophyte moulds. Rev Iberoam Micol. Mar; 20(1): 6-10.
21. Esteban, A.; Abarca, M. L.and Cabañes, F. J. (2005): Comparison of disk
diffusion method and broth microdilution method for antifungal
susceptibility testing of dermatophytes. Medical Mycology 43: 61-66.
22. Faggi, E.; Pini, G.; Campisi, E.; Bertellini, C.; Difonzo, E.; and
Mancianti, F. (2001): Application of PCR to distinguish common species of
dermatophytes. J. Clin. Microbiol. 39: 3382-85.
23.
Favre, B., Hofbauer, B., Hildering, K.S. and Ryder, N.S. (2003): Comparison
of in vitro activities of 17 antifungal drugs against a panel of 20
dermatophytes by using a microdilution assay. J Clin Microbiol. 41: 4817-9
24. Fernindez, T.B., Carrillo, A.J. Martin, E., Del Palacio, A., Moore, M.K.,
Valverde, A., Serrano, M. and Guarro, J. (2001): In vitro activities of 10
antifungal drugs against 508 dermatophyte strains. Antimicrobial Agents and
Chemotherapy 45: 2524-28.
25. Fernindez, T.B.,
Cabanes, F.J.; Carrillo, A.J.; Estaben, A.; Inza, I.; Abarca, L.; and Guarro,
J. (2002): Collaborative evaluation of optimal antifungal susceptibility
testing conditions for dermatophytes. J. Clin. Microbiol. 40:3999-4003.
26. Fisher and Cook (1998): Fundamentals of diagnostic mycology. W.B.
Saunders Company, Philadelphia, London, Toronto, Montreal, Sydney, Tokyo,
p118-155.
27. Friedlander, S.F., Aly, R., Krafchik,
B., Blumer, J., Honig, P., Stewart, D., Lucky, A.W., Gupta, A.K., Babel, D.E.,
Abrams, B., Gourmala, N., Wraith, L. and Paul, C. (2002): Terbinafine in the
treatment of Trichophyton tinea capitis: a randomized, double-blind,
parallel-group, duration-finding study. Pediatrics 109: 602-7.
28. Fuller, L.C., Child, F.C., Midgley, G. and Higgins, E.M. (2003): Scalp
ringworm in south-east London and an analysis of a cohort of patients from a
pediatric dermatology department. Br J Dermatol. 148: 985-8.
29. Graser, Y., El Fari, M., Vilgalys, R., Kuijpers, A.F.A., De Hoog, G.S.
Presber, W. and Tietz, H.J. (1999): Phylogeny and taxonomy of family
Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal
ITS region. Med. Mycol. 37: 105-14.
30. Gupta, A.K.
and Kohli, Y. (2003): In vitro susceptibility testing of ciclopirox,
terbinafine, ketoconazole and itraconazole against dermatophytes and
nondermatophytes, and in vitro evaluation of combination antifungal
activity. Br J Dermatol. 149: 296-305.
31. Gupta,
A.K., Chaudhry, M. and Elewski, B. (2003): Tinea corporis, tinea cruris,
tinea nigra, and piedra. Dermatol Clin. 21:395-400.
32. Hainer, B.L. (2003): Dermatophyte infections. Am. Farm. Physician. 67:
101-08.
33. Howell, S.A., Barnard, R.J. and
Humphreys, F. (1999): Application of molecular typing methods to
dermatophyte species that cause skin and nail infections. J Med Microbiol
48: 33-40.
34. Hubert, J.N. and Callen, J.P. (2003):
Recalcitrant tinea corporis as the presenting manifestation of patch-stage
mycosis fungoides. Cutis. 71: 59-61.
35. Jessup, C.J.,
Warner, J., Isham, N., Hasan, I. and Ghannoum, A.M. (2000): Antifungal
susceptibility testing of dermatophytes: establishing a medium for inducing
conidial growth and evaluation of susceptibility of clinical isolates. J
clin Microbial 38: 341-44.
36. Kano, R., Okabayashi,
K., Nakamura, Y., Ooka, S., Kashima, M., Mizoguchi, M., Watanabe, S. and
Hasegawa, A. (2000): Differences among chitin synthesis 1 gene sequences in
Trichophyton rubrum and T. violaceum. Med. Mycol. 38: 47-50.
37. Kawai, M. (2003): Diagnosis of dermatophytosis: conventional methods and
recent molecular biological methods. Nippon Ishinkin Gakkai Zasshi. 44:
261-4.
38. Koichi, M., Yoshico, T., Takashi, M.,
Atsuhiko, H., Yoshito, T., Ryo, H., Katsuhisa, U., Hiuuuga, S. and Hideyo,
Y. (1999): Phylogenetic classification and species identification of
dermatophyte strains based on DNA sequences of nuclear ribosomal internal
transcribed spacer regions.J Clin Microbiol April; 37: 920-24.
39. Lacroix, C.; Baspeyras, M.; de La Salmoniere, P.; Benderdouche, M.;
Couprie, B.; Accoceberry, I.; Weill, F.X.; Derouin, F. and Feuilhade de
Chauvin, M. (2002): Tinea pedis in European marathon runners. J. Eur. Acad.
Dermatol. Venereol. 16: 139-42.
40. Lehmann, S., Ott,
H., Barker, M., Heimann, G., Poblete-Gutierrez, P. and Frank, J. (2004):
Identification of geophilic and zoophilic dermatophytes in siblings with
tinea capitis a pathogenic factor or contamination? Hautarzt 55:1001-3
(Abstract).
41. Li RY, Li DM , Yu J, Liu W, J ZH and
Wang DL (2004) : Application of molecular biology techniques in the
identification of pathogenic fungi and diagnosis of fungal infection.
Beijing Da Xue Xue Bao; 36 ( 5) : 536 -9.
42. Liu,
D.; Coloe, S.; Baird, R.; and Pederson, J. (1997): Molecular determination
of dermatophyte fungi using the arbitrarily primed polymerase chain
reaction. Br. J. Dermatol. 137: 351-55.
43. Liu, D.,
Coloe, S., Baird, R. and Pedersen, J. (2000a): Rapid mini preparation of
fungal DNA for PCR. J. Clin Microbiol 38:471
44.
Liu, D., Coloe, S., Baird, R. and Pedersen, J. (2000b): Application of PCR
to the identification of dermatophyte fungi. J. Med Microbiol 493:497.
45. Mahmoud, A. (2002): A study of dermatophytosis in Sana´a Yaman Republic.
Mycosis 45: 105.
46. Marie, M.D., Claire, L.,
Martine, F.C., Isabelle, L.G., Catherine, G., Frederic, L. and Francis, D. (
2001):
47. Rapid discrimination among Dermatophytes,
Scytalidium spp., and other fungi with a PCR restriction fragment
48. length polymorphism riboyping method. J. Clin. Microbiol. 39: 685-90.
49. Metin, A.; Subasi, S.; Bazkurt, H.; and Calka, O. (2002): Tinea capitis
in Van, Turkey. Mycoses. 45:492-95.
50. Milne, L.J.R.
(2001): Fungi: In practical medical microbiology. By Mackie and McCartney (eds,),
14th ed Churchill Livingstone, p.695-717
51.
Moubasher,A., Hassan, M.O., Omaima, M.H., Moharram, M.A. and Abeer, S.E.
(2000): Epidemiology of tinea capitis among primary school children in
Assiut city, 1999. (abstract). http://www.aun.eun.eg/fac_med/medicin/april.htm
52. Mounkassa, B., Vandemeulebroucke, E., Jousserand, P. and Poujade, F.
(2004): Tinea capitis occurring in a preschool environment. Ann. Dermatol.
Venereol. 131: 283-4.
53. Mukherjee, P.K.; Leidich,
S.D.; Isham, N.; Leitner, I.; Ryder, N.S. and Ghannoum, M.A. (2003):
Clinical Trichophyton rubrum strain exhibiting primary resistance to
terbinafine. Antimicrob. Agents Chemother. 47:82-86.
54. National Committee for Clinical Laboratory Standards (1997): Reference
method for broth dilution antifungal susceptibility testing of yeasts.
Approved standard M27-A. National Committee for Clinical Laboratory
Standards, Wayne, Pa.
55. National Committee for
Clinical Laboratory Standards (2002): Reference method for broth dilution
antifungal susceptibility testing of filamentous fungi. Approved standard
M38-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
56. Nweze EI and Okafor JI (2005) : Prevelence of dermatophytic fungal
infections in children: a recent study in Anambra. Mycopathologia; 160 (3):
239-43.
57. Pujol, I.; Capilla, I.J.; Ferna´ndez-Torres,
B.; Ortoneda, M.; and Guarro, J. (2005): Use of the Sensititre colorimetric
microdilution panel for antifungal susceptibility testing of dermatophytes.
J. Clin. Microbiol. 40: 2618-22.
58. Omar, A.A.
(2000): Ringworm of the scalp in primary-school children in Alexandria:
infection and carriage. East. Mediterr. Health. J. 6: 961-7.
59. Osborne, C.S.; Hofbauer, B.; Favre, B. and Ryder, N.S. (2003): In vitro
analysis of the ability of Trichophyton rubrum to become resistant to
terbinafine. Antimicrob. Agents and Chemother.47: 3634-36.
60. Riley, L.E. (2004): Laboratory methods used for strain typing of
pathogens: PCR-based strain-typing methods. In Molecular epidemiology of
infectious diseases, principles and practices, American Society of
Microbiology, Washington DC.
61. Seebacher, C.
(2003): The change of dermatophyte spectrum in dermatomycoses. Mycosis. (46)
Suppl 1; 42-6.
62. Shibaki, H. and Shibaki, A.
(2003): Analysis of dermatophyte flora at private clinic in Sapporo during
the period 1992 to 2001. Nippon Ishinkin Gakkai Zasshi. 44: 209-16.
63. Sigurgeirsson, B. and Steingrimsson, O. (2004): Risk factors associated
with onychomycosis. J Eur Acad Dermatol Venereol. 18: 48-51.
64. Sofia, P.; Annette, W.F; Deanna, A.S. and Michael, G.R. (2001):
Comparison of in vitro activities of voriconazole and five established
antifungal agents against different species of dermatophyte using a broth
macrodilution method. J. Clin. Microbiol. 39: 385-88.
65. Summerbell, R.C. (2003): Trichophyton, Microsporum, Epidermophyton, and
agents of superficial mycoses. In Murray, P.R; Baron, E.J.; Pfaller, M.A.;
Jorgensen, J.H.; and Yolken, R.H. (ed.) Manual of clinical microbiology (8th
ed), American Society for Microbiology, Washington, DC p1798-1819.
66. Takahashi, Y. and Nishimura, K. (2002): Dematophyte flora at the
dermatology clinic of Kimitsu Chuo hospital from 1994 through 1999. Nippon
Ishinkin Gakkai Zasshi. 43: 21-7.
67. Tampieri, M.P.
(2004): Actuality on diagnosis of dermatomycosis. Parassitologia. 46: 183-6.
68. Tietz, H.J. (2003): Sweat and public showers. Favorable conditions for
foot fungus in the athelete. MMW Fortschr Med.145:29-30.
© 2006 Egyptian Dermatology Online Journal |


